Integrating Clinical Informatics and Medical Genomics Knowledge for P-Health Care: Visualizing Knowledge Structure and Evolution
Received: 01-Aug-2022, Manuscript No. 74320; Editor assigned: 04-Aug-2022, Pre QC No. 74320; Accepted Date: Aug 01, 2022 ; Reviewed: 18-Aug-2022 QC No. 74320; Revised: 22-Aug-2022, Manuscript No. 74320; Published: 29-Aug-2022, DOI: 10.24105/ejbi.2022.18.8.88-91
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact submissions@ejbi.org
Abstract
Through the use of our expanding knowledge of the molecular causes of disease, genomic medicine seeks to transform the delivery of healthcare. Data sets are big and extremely varied because research in this field is data demanding. Researchers need to integrate these vast and varied data sets in order to derive knowledge from the data. This poses difficult informatic difficulties, including the need to integrate disparate data sources and describe data in a way that is acceptable for computational inference. Bioinformatics and the development of electronic medical records (EMRs) are two important topics in healthcare. The technologies for data collecting, processing, and visualization in two distinct domains were established as a result of the widespread use of EMR systems and the conclusion of the Human Genome (HG-Project). In the not too distant future, personalized, preventive, and predictive healthcare services are anticipated to be made possible by the vast amounts of data from the clinical and biological domains. Four essential informatics domains must be taken into account for the combined use of EMR and bioinformatics data: data modeling, analytics, standards, and safety. Researchers in the fields of clinical informatics and medical genomics will benefit from this study's comprehensive understanding of p-Health care, big data research in healthcare, and future research objectives.
Keywords
p-Health care, EMRs, Human Genome, Genomic Medicine, Clinical informatics
Introduction
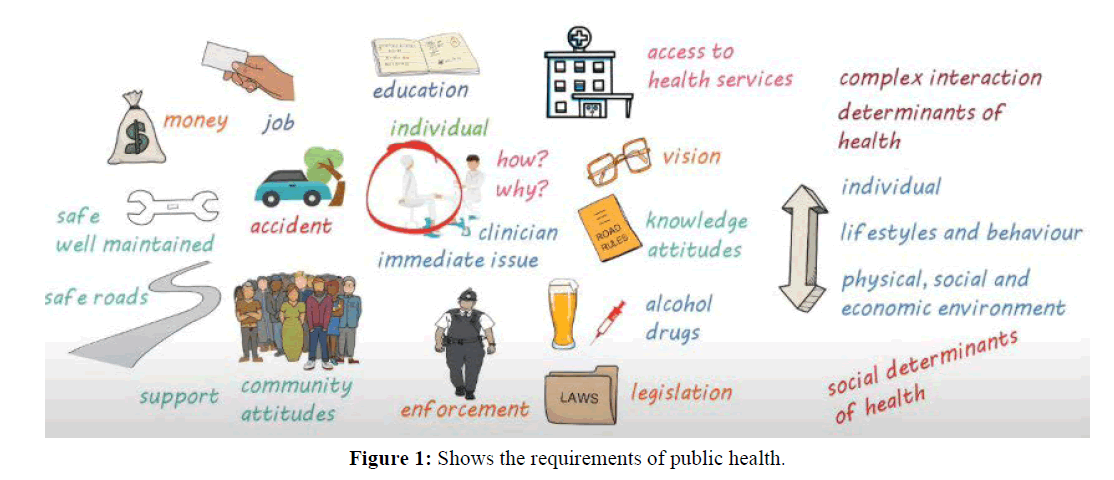
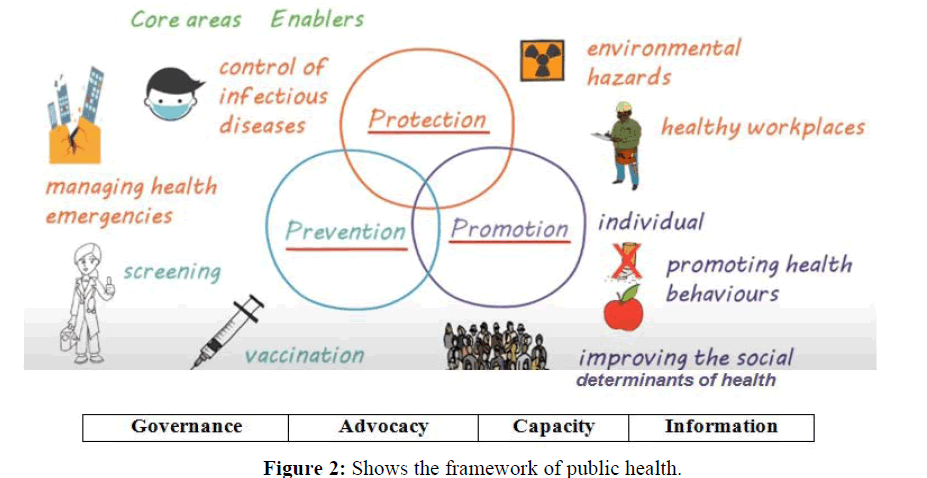
Although the body of research on healthcare big data has expanded quickly in recent years, few studies have utilised bibliometrics and a visualisation strategy to do deep mining and provide an overview of the area. P-Health, which stands for predictive, precise, participatory, preventative, and personalized health, necessitates thorough analyses of each person‘s conditions as recorded by various measurement methods. Requirements and framework are showing in Figures 1 and 2.
Clinical informatics refers to the use of computers to analyse the notes of healthcare professionals and physicians as well as measurement data to enhance the delivery of healthcare. Widespread adoption of electronic health records (EHRs) since the 2010s has significantly advanced clinical informatics development with quickly spreading wearable technologies that continuously record the physiological profile of people both in and outside of the clinic (PHRs or personal health records), supporting m-Health [1,2].
The first electronic digital computers and penicillin, the first antibiotic, were both developed during the 1940s. Due to these ground-breaking developments, some biomedical researchers had begun to investigate the potential applications of digital computers by the late 1950s. The medical sciences, which are by their very nature information-intensive, began to use computers extensively in the 1960s. The necessity for a label for this area of cutting-edge biomedical research and the absence of an English term that encompasses both information and computers led to the invention of the term medical informatics in 1974. The name also required to cover technology, engineering, and science.
In order to ethically incorporate genetic advances into the domains of personalized medicine and public health, public health genomics has developed. A systematic approach is necessary for the appropriate, efficient, and sustainable integration of genetics into healthcare.
Following the promoting health concept, there was a growing understanding that public health had evolved over time and that the government‘s responsibility in delivering public health services required to be outlined, supported, and comprehended
Assessment: determining health issues and priorities while assessing and monitoring the health of vulnerable communities and populations. Information on population health must be regularly and systematically gathered, assembled, analysed, and disseminated in order to achieve this.
Policy Development: Through partnership with stakeholders to develop public policies, programmes, norms, regulations, and resources in order to address prioritised local and national health issues.
Assurance: ensuring that all communities have access to quality care that is both affordable and effective, as well as assessing the success of public health and healthcare initiatives.
Practise of Public Health Genomics
It is obvious how genomics can benefit the practise of public health. But in order to incorporate the advantages of related knowledge and technologies into every facet of providing public health services, it is necessary to take a coordinated approach in order to take advantage of genetic advancements.
To find problems with community health, monitor and evaluate the state of health. Determine and research the community‘s health issues and dangers. People should be informed, educated, and empowered about health issues. Develop community relationships to find and fix health issues. Create plans and policies to aid in promoting the health of the person and the community. Enforce the laws and rules that guarantee safety, preserve the public‘s health and research to find fresh perspectives and creative answers to health issues etc.
Genomic approaches to public health have been successfully incorporated into current paradigms for delivering conventional public health services. Continued integration of genetics and public health holds out the prospect of providing the general public with more specialised, individualised medical care. Governments and policy makers in this field have a special responsibility to steer this effort in a way that ensures the equitable and successful integration of genetic technology and knowledge into health systems [3].
The growth of high-throughput methods and informatics will fundamentally alter numerous fields of medical research and biotechnology. One of the bioinformatics technologies that may be used most easily in clinical medicine and biomedical research is biochip technology. Large-scale profiling of gene expression has shown to be able to categorise certain cancer types. It has also been shown that it is possible to uncover new disease classes and provide prognostic predictions. Changes in molecular cell biology and bioinformatics are revolutionising drug discovery [4].
One example of the great achievements of bioinformatics is the crucial reliance on it for the achievement of the Human Genome Project. The alignment of DNA and protein sequences, genetic variation that occurs naturally, the probability of the structure and function of biomolecules, the analysis of biomolecular interaction networks, the integration of heterogeneous biological databases, the representation of biomolecular knowledge, the simulation of biological processes, the examination of the data produced by large-scale biological experiments, and rational drug design are other areas where bioinformatics has been essential [5].
The majority of researchers concur that the current problem is to comprehend all the data. Data generation now happens more quickly than interpretation. The development of biochips that track the functional activity of genes and proteins has made this situation much more critical.
Big Data is defined by three factors: volume, diversity, and velocity. These traits collectively characterise „Big Data.“ Healthcare experts and academics can forecast public health trends by examining these health records, including how patients may respond to particular therapies and the likelihood of upcoming diseases and pandemics [5]. Big Data Analytics can shed light on clinical data and help decision-makers make well-informed choices on patient diagnosis and care, illness prevention, and other issues. By utilising the possibilities of the data, big data analytics can also increase the effectiveness of healthcare organisations. Applications of big data analytics can enhance patient-based care, detect diseases early, produce new knowledge about how diseases work, keep track of the standard of medical and healthcare institutions, and give better treatment overall [6].
The Serious Issues with Healthcare Data Security at the Present EHRs and the exchange of health information. Adoption of technological mistakes by users. The use of mobile and cloud technology in healthcare technology that is out of date. Health care industry advantages of big data and big data analytics: less medical mistakes, mass illness prevention, preventative care, modeling the spread of diseases, early disease detection, more precise treatment, real-time alerting, patient-specific care, etc. Big Data and the Future of Healthcare; Sector players, including healthcare providers, payers, and insurers, are using the enormous influx of data in healthcare to alter the industry by promoting the best patient care outcomes and lowering healthcare costs at the same time.
Genomic sequencing is quickly advancing into clinical practise, and over $4 billion in significant government funding has been invested in healthcare systems worldwide, in at least 14 different nations. These national genomic-medicine efforts are solving implementation challenges and accumulating data for wider acceptance while simultaneously bringing about dramatic change in the real world.
It is now possible to implement genomically-informed personalized cancer therapy as a route to precision oncology thanks to the rising affordability of next-generation sequencing. Clinicians must interpret the patient‘s genetic abnormalities and choose the best authorized or exploratory medicine, which is extremely difficult due to the complexity of genomic information. To assist clinical decisions and quickly test clinical hypotheses, a sophisticated and useful information system is urgently needed [7].
Numerous studies using extensive health record datasets have been carried out in a variety of societies worldwide. For instance, extensive data gathering and analysis are part of the NSF BIGDATA programme solicitation, which is partially sponsored by the National Institutes of Health (NIH). By managing, analysing, visualizing, and extracting useful data from massive, varied, dispersed, and heterogeneous datasets, this programme seeks to advance fundamental scientific and technological methods.
The Nationwide Human Genome Research Institute (NHGRI) established the Electronic Medical Records and Genomics (eMERGE) Network as a national partnership to create, share, and implement research methods. It combines DNA biorepositories with EMR systems for large-scale, high-throughput genetic studies with the eventual goal of providing genomic testing findings to patients in a healthcare context. The network is currently investigating more than a dozen phenotypes. At sites throughout the network, a number of models for returning clinical results have been put into place or are slated for pilot. The topics of bioinformatics, genetic medicine, privacy, and community involvement are particularly pertinent to eMERGE [8].
Issues with Genome-Enabled EMRs
The main difficulty is combining the diverse data into a single database system. Should there be a single database or should a federated approach be taken into consideration? Additionally, different cases by different users should be taken into account because this would affect the overall system architecture. Moving massive amounts of genomic data is not possible. What should the management strategy and location for high-intensity computing be then? A single person‘s projected raw sequencing data volume is in the neighbourhood of 4 terabytes. The prevention, diagnosis, and treatment of disease may be impacted by the integrated database. It‘s crucial to link clinical data with genetic data in order to fulfill this aim. There will be more clinical applications of genetics for disease diagnosis and treatment planning.
Researchers have examined the opinions and/or understanding of various health professionals about genetics and/or genomics. To the best of our knowledge, however, this is the first investigation into public health educators‘ views, awareness, and understanding of PHG-related genomics. Public health educators play a crucial role in genomics because they work closely with other healthcare professionals to promote the public‘s ability to make informed decisions, increase health literacy, and support the maintenance of healthy behaviour. They provide a variety of educational and health promotion services to the public and collaborate closely with other healthcare professionals to do so [9].
Our sample‘s knowledge of genomics was shown to have a nontrivial knowledge gap, according to the findings. Given the topic of the survey, it‘s possible that participants with more indepth knowledge of genomics chose to participate in this study, making it possible that public health educators‘ understanding of the subject is less comprehensive than what we found. Additionally, we saw that responders performed better on „applied“ knowledge questions than „basic“ ones. It makes natural that basic genomics could not be appreciated as highly as applied genomics given that public health education is an applied professional discipline [10]. Public health educators should be trained in fundamental genomics concepts and methods in order to prevent them from misrepresenting genomic information to the public and obstructing the practise of genomics-related health promotion. A „push“ for incorporating genomic competencies without the requisite background knowledge should be wisely avoided. We anticipate that six critical areas—data sharing, interoperability, equitable access, consumer empowerment, behavior change, and scientific advancement—will work together to transform the current health system from diagnosis reactionary care to preventative measures and well-being in the future of health.
Conclusion
The development of personalized predictive medicine may be aided by genomic information, which has the potential to enhance the detection and treatment of genetic and complex diseases, including cancer. Large amounts of genomic data and difficult computational hurdles have been produced by the greater throughput and quickly declining costs of next-generation sequencing. Accordingly, the transition from this potent finding research to clinical implementation can only be made through careful integration with EMRs, a tool used for front-line patient care. The most important justification for uniting clinical and biological data under one system is to enable cross-disciplinary interchange of information in both directions between two fields with diverse histories and cultures. Additionally, open worldwide cooperation will offer chances to advance the knowledge about, access to, and prevention of human diseases.
References
- Khoury MJ, Bowen MS, Clyne M, Dotson WD, Gwinn ML, Green RF, et al. From public health genomics to precision public health: a 20-year journey. Genetics Med. 2018; 20(6):574-582.
- Hersh WR. The electronic medical record: Promises and problems. J Ame Society Info Sci. 1995; 46(10):772-776.
- Molster CM, Bowman FL, Bilkey GA, Cho AS, Burns BL, Nowak KJ, et al. The evolution of public health genomics: exploring its past, present, and future. Frontiers Pub Health. 201; 6:247.
- Kim JH. Bioinformatics and genomic medicine. Genetics Med. 2002; 4(6):62-65.
- Louie B, Mork P, Martin-Sanchez F, Halevy A, Tarczy-Hornoch P. Data integration and genomic medicine. J Biomed Informatics. 2007; 40(1):5-16.
- Elgendy N, Elragal A. Big data analytics: a literature review paper. In Industrial conference on data mining 2014; 214-227.
- Jang Y, Choi T, Kim J, Park J, Seo J, Kim S, et al. An integrated clinical and genomic information system for cancer precision medicine. BMC Medical Genomics. 2018; 11(2):95-103.
- Choi IY, Kim TM, Kim MS, Mun SK, Chung YJ. Perspectives on clinical informatics: integrating large-scale clinical, genomic, and health information for clinical care. Genomics Informatics. 2013; 11(4):186.
- Chen LS, Goodson P. Public health genomics knowledge and attitudes: a survey of public health educators in the United States. Genetics Medicine. 2007; 9(8):496-503.
- Wiley K, Findley L, Goldrich M, Rakhra-Burris TK, Stevens A, Williams P, et al. A research agenda to support the development and implementation of genomics-based clinical informatics tools and resources. J Ame Med Info Ass. 2022.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref