Cluster Analysis of Rabies Virus-Host (Homosapiens) Network to Determine Various Viral Infections
P. Jaya Simha Reddy, Andhra Loyola College, India, Email: Jayasimhar200@gamil.com
Received: 07-Apr-2021 Accepted Date: May 05, 2021 ; Published: 12-May-2021
Citation: Sai Sampath Y, Jaya Simha P. (2021). Cluster Analysis of Rabies Virus-Host (Homosapiens) Network to Determine Various Viral Infections. EJBI. 17(5)
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact submissions@ejbi.org
Abstract
Rabies is also known as Rhabdoviruses. It is a preventable viral disease transmitted through the bite of a rabid animal. It belongs to the family Rhabdoviridae order mononegavirals. Rabies infects the CNS of mammals, ultimately causing disease in the brain and death. Rhabdoviruse is approximately 180nm long, 70nm wide and 400 trimeric spikes which are tightly arranged on the surface of the virus. The genome encodes 5 proteins designated as nucleoprotein(N), phosphoprotein(P), matrix protein(M) ,glycoprotein(G) and a viral RNA polymerase protein(L). The clinical course of rabies occurs in five stages. Stage 1 is the incubation period, stage 2 is the prodromal period, stage 3 is the neurological period, stage 4 is coma stage 5 occurs infrequently is recovery. The current study determines the biological process of a virus against(Homosapiens). Here we retrieved an interaction network of rabies virus-host(homosapiens) from a string virus database. This network was further be analyzed and the resulted network was clustered. By performing gene ontology analysis for the clustered proteins we, therefore, identified proteins which has a highly effective role in cellular processes and viral infection mechanisms. Hence this study helps to understand the various proteins that can be targeted for further development in drug discovery and also in the prevention of this disease.
Keywords
Rhabdovirus; Monogavirals; CNS; Prodromal; Neurological; Coma; Homosapiens
Introduction
Rabies is a notifiable pathogen. It belongs to order mononegavirales, a virus with a nonsegmented, negativestranded RNA genome. Bullet or rod distinct shapes are classified in the Rhabdoviriade family, these include at least three genera of animal virus lyssavirus, Ephemerovirus, and vesiculovirus. The Centre for Disease Control and Prevention (CDC) reported every year occurs in wild animals like bats, raccoons, skunks, and foxes although every mammal can get rabies [1].
The length of genomic rabies virus encodes 5 proteins including nucleoprotein(N), phosphoprotein(P), matrix protein(M) ,glycoprotein(G) and a viral RNA polymerase protein(L). The mortality rate of neglected viruses leads to death once the symptoms develop and a mortality rate of 1:1,00,000 to 1:1,000 per year. Inducing severe immune responses are deceased intriguingly display know neural damage, neuro histopathological evidence. The virus travels from the muscle tissue to the nervous system, migrates to the spinal cord, and freely covers certain parts of the brain, in an organized high jacking program subsequently to the next host the other organs centrifugally spread the virus. The first defense line against viral infection, although the host innate immune response including TLR, type 1 interferon, TNF alpha, and IL-6, and this virus easily progress in the nervous system. RABB suggests that this has a specific mechanism to suppress host innate immunity. When the virus is injected intracerebrally in high dose to several laboratory strains of the RABB in addition to the wild types causes vital acute encephalomyelitis associated with inflammation of the spinal cord and brain which leads to coma and death [2]. World wide more than 29M people receive a positive bite on vaccination every year. To prevent hundreds of thousands of rabies deaths annually have been estimated. Rabies is estimated that US$ 8.6 billion per year as the economic burden of dog-mediated rabies globally. Histopathological changes indicative of apoptosis are necrosis in infected cells that don‘t include constructive attenuated strains, while type strains and CVS-11. Rabies encephalitis remains a mystery due to over 100 years of controlling rabies by developing RABB vaccines and sclerotherapy, precise neurological and immunological etiology as well as rare survival cases.
For a better understanding of rabies fatal mechanism, some students have started after the emergence of omics technology they pave the way towards rabies fatal mechanism. Rabies progression has been based mainly on analyzing gene expression alterations in elucidating the sessional biological processes. Profiling of m-RNA and micro RNA of a rabies-infected cell has been reported by Zhao et al. [2] . The gene expression profile of CNS tissue infected with CVS-11 has been analyzed by Sugiura et al. [2]. Neurons infection with recombinant RABB expressing CRE-recombinase which changes in gene expression were also studied in marked. In different species, numerous other studies have also analyzed gene expression profiling using transcriptomic are proteomic methods with the diverse cellular model.
To increase the reliability of results and generalizability of this independent but related study‘s, it is recommended to statically combine such data commonly known as data integration metaanalysis including infectious disease several studies have shown the benefits of meta-analysis in terms of both higher statistical power and precision in detecting deferentially expressed gene (DEGs) in different complex traits. To improve the representation of data, data integration approaches at a higher level try to map multi biological data levels into one mechanistic network. Regardless of inter studies differences the generated multidimensional network is likely more useful inferring universally involved processes or pathways.
We horizontally integrated nine high-throughput transcriptome databases to identify consensus DEGs, having in mind the common concerns in meta-analysis. Based on protein-protein interaction (PPIN) and signaling pathways by defining the identified DEGs as seed genes which under laying molecular network in rabies pathogenesis was then extracted. We experimentally several key DEGs in rabies-infected cells by using real-time PCR. Based on integrated omics data sets and experimental validation, may be used to shed like on a vague portrait of complex disease pathobiology which we demonstrated that system biomedicine approached [2].
Materials and Methods
4.1 Collection of data set
For having a better understanding of virus-host protein-protein interactions the development of treatments and vaccines as viruses continue to pose risks to global health. A protein-protein interaction database specifically catering to virus-virus and virushost interaction, through here we introduce STRING VIRUS DATABASE (https://string-db.org). From experimental and text mining channels to provide combine possibility for interaction between viral and host proteins which combines the evidence of this database. Between 239 virus 319 hosts, there are 1,77,425 interactions are there in this string virus database. The interaction data can be accessed to the latest version of the Cytoscape STRING app and this database is available for viruses. The result are listed as four distinguished categories of highest confidence score(0.9-1), high confidence (0.7-0.8), medium confidence (0.4- 0.6) and low confidence(up to 0.1). The confidence score is an indicator of interactions between nodes that are connected by multiple paths. String-db.org [3].
4.2 Network analysis
For integrating biomolecular interaction networks with high throughput expression data and other molecular states into a unified conceptual framework Cytoscape (http://www.cytoscape. org/ ) is an open-source software project. Cytoscape is most powerful when used in connection with a large database of protein-protein, protein DNA and genetic interactions that are increasingly available for human and model organisms although applicable to any system for molecular components and interactions. To visually integrate the network with expression profiles phenotype and other molecular states and to link the network to databases of functional annotations, basic functionality to layout and query the networks Cytoscape‘s software core provided. Through a straightforward plugin architecture, allowing rapid development of additional computational analysis and features these core is extensible. For a search of interaction pathways core relating with changes in gene expression, a study of protein complex s involved in cellular recovery to DNA damage which interference of a combined physical/functional interaction network for Halobacterium and interphase to detailed stochastic/ kinetic gene regulatory models for these Cytoscape plugins are surveyed [4].
4.3 Topological analysis
Various experimental techniques and computational prediction methods are rapidly increasing in amounts of molecular interaction data are being produced. We have developed the versatile Cytoscape plugin network analyzer to gain insight into the organization and structure of the resultant large complex networks are formed by interacting molecules. The number of nodes, edges, and connected components, the network diameter, radius, density centralization, heterogeneity and clustering coefficient, the characteristic path length & distributions of node degrees, neighborhood connectives, average clustering coefficient, and shortest path length which computes and displays a comprehensive set of topological parameters. To construct the intersection are the union of two networks that contains extra functional both direct and indirect networks can be applied from network analyzers. From the user an interactive and high customizable application that requires no expert knowledge from graph theory [5].
4.4 Clustering analysis
Cytoscape was comprised of a plugin called Mcode. This was used to identify the densely connected node from a highly dense network. This helpful in separating biological modules are related set of nodes. The IAV/human protein/protein interaction was analyzed using the plugin to construct highly interconnecting clusters [6]. For our analysis, we set the parameters as node score cutoff: 0.2; haircut: true, fluff: false, K-core: 4, and max depth from seed: 100.
4.5 GO analysis
A resource for the evolutionary and functional classification of genes from organisms across the tree of life is known as PANTHER (Protein Analysis Through Evolutionary Relationships). For the past 2 years, we report the improvements that we have made to the resource. We have added prokaryotic and plant genomics to the phylogenetic gene trees, expanding the representation of gene evolution in these lineages, for evolutionary classifications. The MEROPS resource for protease and protease inhibitory families which have refined many protein‘s family boundaries and has a lined Panther. We have developed an entirely new PANTHER GOslim containing over 4 times as many Gene Ontology terms as our previous GO-slim, as well as the curated association of genes to these terms, for functional classifications. Panther website: users can now analyze over 900 different genomes, using updated statistical tests with false discovery rate corrections per multiple testing at lastly we have made substantial improvements to the enrichment analysis tools available. For easy addition to the third party cites the overrepresentation test is also available as a web service [7].
Results
5.1 Protein-protein network analysis
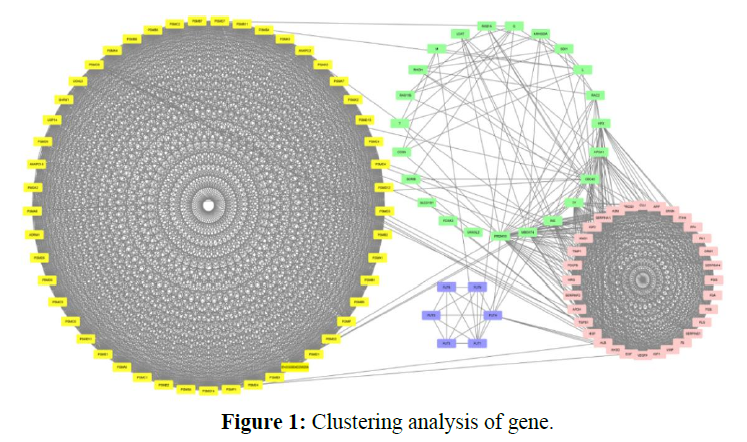
This is the study of protein-protein interaction of rabies virus. We developed a spatially explicit network construction to expand the network and nodes by using the String virus database. In the string virus database medium confidence, 0.400 is the minimum required score for interaction and a maximum number of interactions of 1st shell 200 and 2nd shell 100. The network is updated and exported. Thus a network of 1884 interactions resulted. The network is reconstructed with Cytoscape which occurred in the network of 110 nodes and 1884 interactions, and these interactions were shown in the network Figure 1.
Gene clusters as viewed in Cytoscape as shown in the figure along with unclustered genes. The clusters are ranked from C1-C3 according to Mcode score. For easy distinction, genes belonging to cluster 1 are highlighted with yellow color, cluster 2 is highlighted with pink color, cluster 3 is highlighted with blue color, and unclustered genes are highlighted with light green color.
5.2 The Clustering analysis
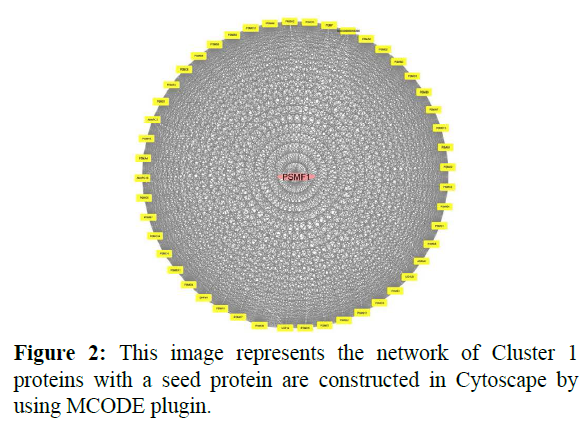
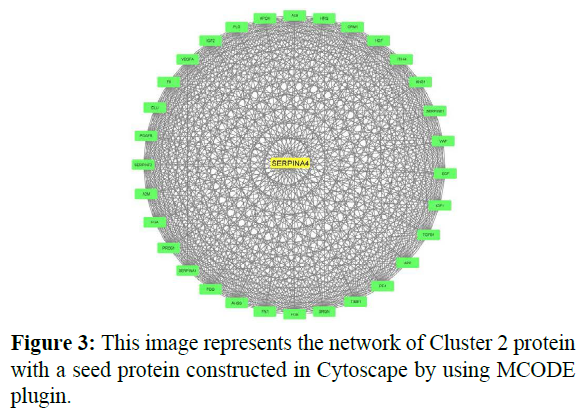
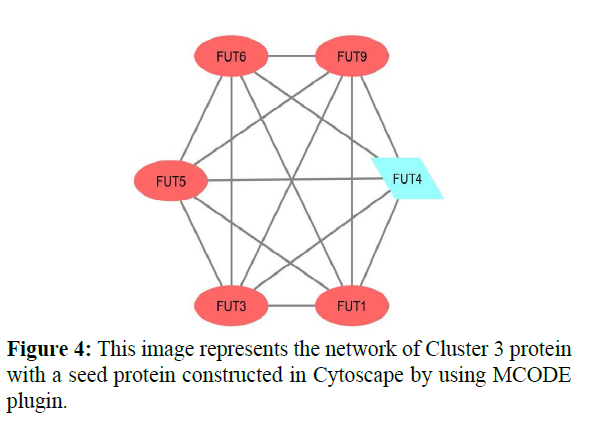
In cluster analysis, the network was analyzed using MCODE plugin which resulted in 3 highly clustering (C1-C3) which has total nodes 110. Through this 3 clusters (C1, C2, C3) were identified in gene expression in various processes and functions. Cluster 1 has a high number of interactions score 1150 with 49 nodes, in this, the seed protein is PSMF1 as shown in Figure 2. Cluster2 was the second-highest interaction score 496 with 32 nodes including a seed protein SERPINA4 as shown in Figure 3. Cluster 3 has the least interaction score of 6 with 15 nodes including a seed protein FUT4 respectively as shown in Figure 4.
5.3 Gene ontology
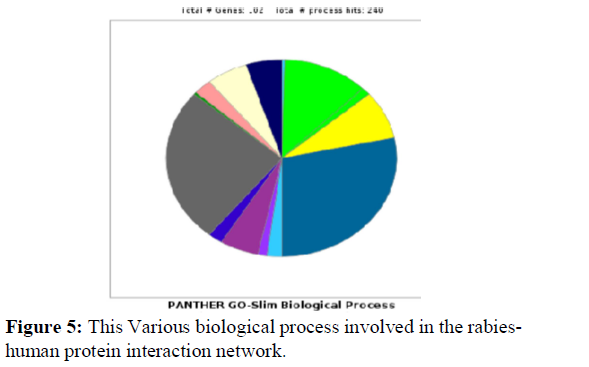
Gene Ontology-based analysis is done for the resulted clustered proteins to identify their various biological processes by using the panther GO database. Here the interaction network of Rabieshuman proteins was analyzed for 87 Clustered proteins for their respective GO terms biological process. Further, these proteins were analyzed for GO terms cellular processes (GO:0009987) and about 64 proteins were identified and these proteins were further analyzed for various Go terms involved in cellular processes as shown in Figure 5 and Table 1.
| SlNo | Biological process | GO terms | No.of proteins |
|---|---|---|---|
| 1 | response to stimulus | (GO:0050896) | 1 |
| 2 | signaling | (GO:0023052) | 1 |
| 3 | cellular process | (GO:0009987) | 42 |
| 4 | metabolic process | (GO:0008152) | 41 |
| 5 | biological regulation | (GO:0065007) | 7 |
| 6 | cellular component organization or biogenesis | (GO:0071840) | 11 |
| 7 | cellular component organization or biogenesis | (GO:0071840) | 1 |
| 8 | cellular process | (GO:0009987) | 14 |
| 9 | multi-organism process | (GO:0051704) | 1 |
| 10 | localization | (GO:0051179) | 6 |
| 11 | biological regulation | (GO:0065007) | 13 |
| 12 | response to stimulus | (GO:0050896) | 7 |
| 13 | signaling | (GO:0023052) | 5 |
| 14 | developmental process | (GO:0032502) | 1 |
| 15 | multicellular organismal process | (GO:0032501) | 2 |
| 16 | locomotion | (GO:0040011) | 3 |
| 17 | metabolic process | (GO:0008152) | 11 |
| 18 | cell population proliferation | (GO:0008283) | 3 |
| 19 | immune system process | (GO:0002376) | 2 |
| 20 | cellular process | (GO:0009987) | 1 |
| 21 | metabolic process | (GO:0008152) | 6 |
Table 1: Biological processes.
The Cluster 1 was subjected for gene enrichment analysis where 56 proteins were isolated which functions for various cellular processes like cell cycle process(GO:0022402), cell cycle (GO:0007049), cellular component (GO:0016043), cellular metabolic process (GO:0044237), cell communications (GO:0007154) and Cellular response to stimulus (GO:0051716) only one protein is involved. 2 proteins were involved in cell cycle process (GO:0022402) and cell cycle (GO:0007049). 11 proteins are involved in Cellular Component (GO:0016043). 41 proteins are involved in Cellular Metabolic process (GO:0044237). The proteins are involved in cellular functions as shown in Table 2.
| Cellular processes (GO:0009987) of Cluster 1 | |
|---|---|
| Mapped I’d | Functions |
| PSMC5 | Cell Communications (GO:0007154) |
| PSME2, PSME1 | Cell cycle process (GO:0022402) Cell Cycle (GO:0007049) |
| PSMC4, PSMC6, PSMD11, PSMC3, PSME2, PSMD13, PSME1, PSMC2, POMP, PSMD4, PSMC5 | Cellular Component (GO:0016043) |
| PSMC6 | Cellular response to a stimulus (GO:0051716) |
| PSMC4,PSMB8,PSMC6, PSMB11,PSMD6,UCHL5, PSMD14,PSMA4,PSMA3, PSMD2,PSMD11,PSMD8, PSMC3,PSMA7,PSME2, PSMB1,PSMA7,PSMD7, PSMB6,PSMB5,PSMD13, PSMA5,ANAPC16,PSME4, PSMA6,PSMB3,PSME1, PSMC2,PSMD12,PSMA2, PSMP1,PSMF1,PSMB4, PSMB7,PSMA8,ADRM1, PSMD3,PSMB9,PSMB2, PSMD4, PSMC5 | Cellular Metabolic process (GO:0044237) |
Table 2: List of Genes shows various functions in Cluster 1 proteins.
As the above cluster, Cluster 2 was also analyzed for gene enrichment analysis where 22 proteins were isolated which functions for various cellular processes. Only one protein has involved in Cell activation (GO:0001775), Cellular component organization (GO:0016043), Microtubule process (GO:0007017), Vesicle targeting (GO:0006903). 5 proteins are involved in Cell Communication (GO:0007154), Cellular response to a stimulus (GO:0051716), Signal transduction (GO:0007165). 4 proteins are involved in the Movement of the cell (GO:0006928). 11 proteins are involved in the Cellular metabolic process (GO:0044237). The proteins are involved in cellular functions as shown in Table 3.
| Cellular processes (GO:0009987) of Cluster 2 | |
|---|---|
| Mapped I’d | Functions |
| HRG | Cell activation (GO:0001775) |
| TGFB1, PDGFB, VEGFA, PF4, EGF | Cell Communication (GO:0007154) Cellular response to a stimulus (GO:0051716) Signal transduction (GO:0007165) |
| ALB | Cellular component organization (GO:0016043) Microtubule process (GO:0007017) Vesicle targeting (GO:0006903) |
| TGFB1, KNG1, SERPINE1, SERPINE2, PDGFB, TIMP, SERPINE4, SERPINA1, VEGFA, AHSG, HRG | Cellular metabolic process (GO:0044237) |
| PDGFB, VEGFA, ALB, PF4 | Movement of the cell (GO:0006928) |
Table 3: List of Genes shows various functions in Cluster 2 proteins.
Finally, cluster 3 was analyzed for gene enrichment analysis where 7 proteins were isolated which functions for various cellular processes. Only one protein has involved in the Biosynthetic process (GO:0009058), Primary metabolic process (GO:0044238), Cellular metabolic process (GO:0044237), Nitrogen compound metabolic process (GO:0006807), Organic substance metabolic process (GO:0071704). 6 proteins are involved in Glycosylatin (GO:0070085) (Table 4).
| Cellular processes (GO:0009987) of Cluster 3 | |
|---|---|
| Mapped I’d | Functions |
| FUT1 | Biosynthetic process (GO:0009058) Cellular metabolic process (GO:0044237) Primary metabolic process (GO:0044238) Nitrogen compound metabolic process (GO:0006807) Organic substance metabolic process (GO:0071704) |
| FUT9, FUT6, FUT5, FUT3, FUT4, FUT1 | Glycosylation (GO:0070085) |
Table 4: List of Genes shows various functions in Cluster 3 proteins.
About 64 proteins were identified involving various cellular processes, these proteins were cross-checked in the UniProt database for viral infection mechanism which resulted in giving 15 proteins out of 64 which are involved in viral infection mechanisms. The 15 proteins which cause viral infection are PSMB8, PSMA4, PSMA3, PSMC3, PSMB1, PSMA7, PSMB6, PSMB5,PSMB3, PSMA2, PSMB4, PSMB7, SERPINE2, PSMB9, PSMB2 as shown in the Table .5.
| Protein Id | Biological Function |
|---|---|
| PSMB8,PSMA4,PSMA3,PSMC3,PSMB1,PSMA7,PSMB6,PSMB5,PSMB3,PSMB4,PSMB7,PSMB9,PSMB2,SERPINE2 | Viral process |
| PSMA2 | Response to virus |
Table 5: Various protein involved in viral infection and mechanism.
Discussion
The oldest disease known for mankind is the rabies virus. The pathogenic mechanism of the rabies virus leads to the development of neurological disease and death is still yet poorly understand [8]. RABV surface protein has characterized and was observed that anti-RABV-G monoclonal antibodies protected laboratory animals for the RABV challenge. The pathogenic and apathogenic counterpart strains have continued to enable investigations into RABV-G as a molecular determinant of pathogenicity. In this manner, one mutant has discovered which substitutes RABV-G‘s position 333 arginine with glutamate or isoleucine, has become a well-studied model of G-RABV attenuation. The G-333 mutant displays spreading, infecting primary motoneurons following the intramuscular inoculation becomes blocked after the first cycle of infection. For instance, the RABV-G of a fixed, nonpathogenic strain of RABV(SN-10) with a RABV-G of a bat-associated street virus (SHBRV) or that of two different fixed pathogenic strains (CVS-N2c and CVS-B2c). This resulted in the incomplete, restoration of the pathogenic after intramuscular inoculation for each of chimeric viruses [9].
The rabies virus vaccines have been existing for decades, each year rabies virus infections can still cause around 50,000 people were infected worldwide. We can see most of the cases occurs in developing countries, where these vaccines are not available [10]. 40% of people bitten by suspect rabid animals are under 15 years of age. Reason for this because high costs of cell culture or egggrown rabies virus vaccines and lack of functional cold chains in many regions in which rabies virus is endemic. This vaccine should be stored at -80c to up to +70c for several months did not impact the protective capacity of the mRNA vaccine. One dose of reconstituted rabies vaccine contains less than 100mg human albumin, less than 150mcg neomycin sulfate, and 20mcg of phenol red indicator. Beta-propiolactone, a residual component of the manufacturing process, is present in less than 50 parts per million.
In this experiment, we integrate the protein interaction data by different computational methods and various databases to obtain highly rabies-associated host proteins to provide information and vaccine for the virus. Here we used Cytoscape as a platform and its important plugins and other tools to explore the Rabieshuman protein interactions and found 110 host proteins that are associated with rabies, among them about 15 proteins are responsible for various rabies infection mechanisms and can be taken as a potential drug target for the cure of rabies infection. The data we structured in this study was taken from a vast variety of literature sources and various databases. Therefore the constructed rabies-human protein interaction network and identified proteins can be benefited from various drugs.
CONCLUSION
Rabies virus was caused by lyssavirus which is a worldwide problem with seasonal and pandemic characteristics. These days Scientists are using computational biology to investigate deep into the host factors responsible for viral infection. Our work concentrates on proteins involved in the rabies infection mechanism in humans and was explained by computational work.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
The authors would like to thank Dr. Gollapalli Pavan for his help in the initial phase of research work. The author would like to thank the management of NITTE for providing the necessary facilities to carry out this project work.
REFERENCES
- http://www.cdc.gov/
- Jamalkandi SA, Mozhgani SH, Pourbadie GH, et al. Systems Biomedicine of Rabies Delineates the Affected Signaling Pathways. Front Microbiol. 2016; 7:1688.
- Cook HV, Doncheva NT, Szklarczyk D, von Mering C, Jensen LJ. Viruses. STRING: A Virus-Host Protein-Protein Interaction Database. Viruses. 2018; 10(10):519.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11):2498-2504.
- Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008; 24(2):282-284.
- Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014; 47(8).13-24.
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim, and improvements in enrichment analysis tools. Nucleic Acids Res. 2019; 47(D1):D419-D426.
- Mehta SM, Banerjee SM, Chowdhary AS. Postgenomics biomarkers for rabies—the next decade of proteomics. OMICS. 2015; 19(2):67-79.
- Davis BM, Rall GF, Schnell MJ. Everything You Always Wanted to Know About Rabies Virus (But Were Afraid to Ask). Annu Rev Virol. 2015; 2(1):451-471.
- Stitz L, Vogel A, Schnee M, et al. A thermostable messenger RNA based vaccine against rabies. PLoS Negl Trop Dis. 2017; 11(12):e0006108.