Analysis Unit Data Model for Statistical and Machine Learning Analysis using the Health Insurance Claims Database
Received: 29-Jul-2022, Manuscript No. 72193; Editor assigned: 01-Aug-2022, Pre QC No. 72193; Accepted Date: Aug 29, 2022 ; Reviewed: 15-Aug-2022 QC No. 72193; Revised: 18-Aug-2022, Manuscript No. 72193; Published: 25-Aug-2022, DOI: 10.24105/ejbi.2022.18.8.80-87
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact submissions@ejbi.org
Abstract
Background: Administrative databases of health insurance claims are becoming increasingly popular. However, since they generally contain only the data necessary to assess claims, they are insufficient for research purposes, and the data are normalized in such a manner that patient-care data are dispersed across multiple tables. Thus, creating a dataset that is appropriate for analysis requires a great deal of effort and involves techniques that would be difficult for clinicians. Objectives: The aim of the present study was to create a data warehouse (DW) that could provide easy access to the data required for epidemiological research Methods: First, epidemiological studies that used data from the National Database of Health Insurance Claims and Specific Health Checkups of Japan (NDB) as a source were surveyed to identify the attributes (variables) most commonly analyzed. Subsequently, these attributes were extracted from the NDB to construct a data model suitable for the analysis of single-patient units. Results: A DW featuring attributes frequently used in epidemiological research, which are also integrated at the per-patient level, was constructed. The DW was then used in two studies: one concerning postpartum hemorrhage and one concerning patients after cardiopulmonary resuscitation. Consequently, four of the six types (approximately 67%) and four of the seven types (approximately 57%) of the required attributes were available through the DW. Conclusion: This study constructed a DW by rendering the attributes that are frequently used in epidemiological analyses. This represents the first step in building a common infrastructure based on the NDB.
Keywords
Data warehouse, Epidemiology, Health insurance claims Database, NDB, Relational model
Introduction
In Japan, the importance of applying health and medical care databases, such as databases of health insurance claims, is beginning to be recognized, and there is a flourishing new body of epidemiological studies that use these databases as source material. However, the application of these data is not maximized because of two main challenges: the quality of the data and the structure of the databases. Considering the former, because the primary purpose of these databases is health insurance claims and registering patient data, data that are commonly required for epidemiological studies may not be stored. Thus, researchers must create their own markers or indices, or process the data themselves, which is burdensome. With regard to the second challenge, the majority of the databases currently in use are relational databases (RDBs), which are designed to support the relational model [1] created based on the theory of sets and logic. In the relational model, data are distributed across multiple tables using a logic circuit design that aims to normalize redundant data and increase the efficiency of data storage and updates [2]. RDBs have been used in several countries to build databases for health-insurance claims [3]; however, in general, a per-patient data structure is considered best for clinical or epidemiological research [4]. Thus, researchers who wish to use RDB-based databases as a source material must manipulate the database for analysis, and extract and integrate data for each patient from the multiple tables in which they are dispersed. This inconvenience has inspired several studies to attempt to reconstitute items in such databases into per-patient tables, which are more suitable for statistical analysis [5]; however, large obstacles still remain for users who are relatively unskilled and/or inexperienced in database manipulation. Because this secondary use of health insurance claims databases has a relatively short history, users who can overcome the two heterogeneous problems are a minority. To promote more active use and application of these databases, it is necessary to resolve as many of these problems as possible far in advance.
The primary aim of this study was to solve the two abovementioned problems by identifying items that are frequently used in epidemiological analysis, through a review of previous articles that have used the National Database of Health Insurance Claims and Specific Health Checkups of Japan (NDB), and by supplementing as many missing items as possible by introducing external master data or by modifying existing data. The secondary aim was to construct an optimal data model (corresponding to “dataset” in epidemiological terminology) for statistical analysis by integrating items identified as being frequently used, which are currently dispersed through multiple tables, into a concentrated per-patient table. We then apply this “data warehouse” (DW) in epidemiological studies conducted by the Obstetrics and Gynecology Department and the Primary Care and Emergency Medicine Department of Kyoto University Hospital, which serves to assess how comprehensively the DW provides the items necessary for writing an epidemiology-based article.
Materials and Methods
Materials
The NDB was created in FY2009, and operated by the Japanese Ministry of Health, Labor, and Welfare (MHLW). To date, there are over 15 billion entries of electronic data concerning health insurance claims and specific health checkups. Secondary use of such data is possible; however, as mentioned in the previous section, their use in research and public surveys is limited [6].
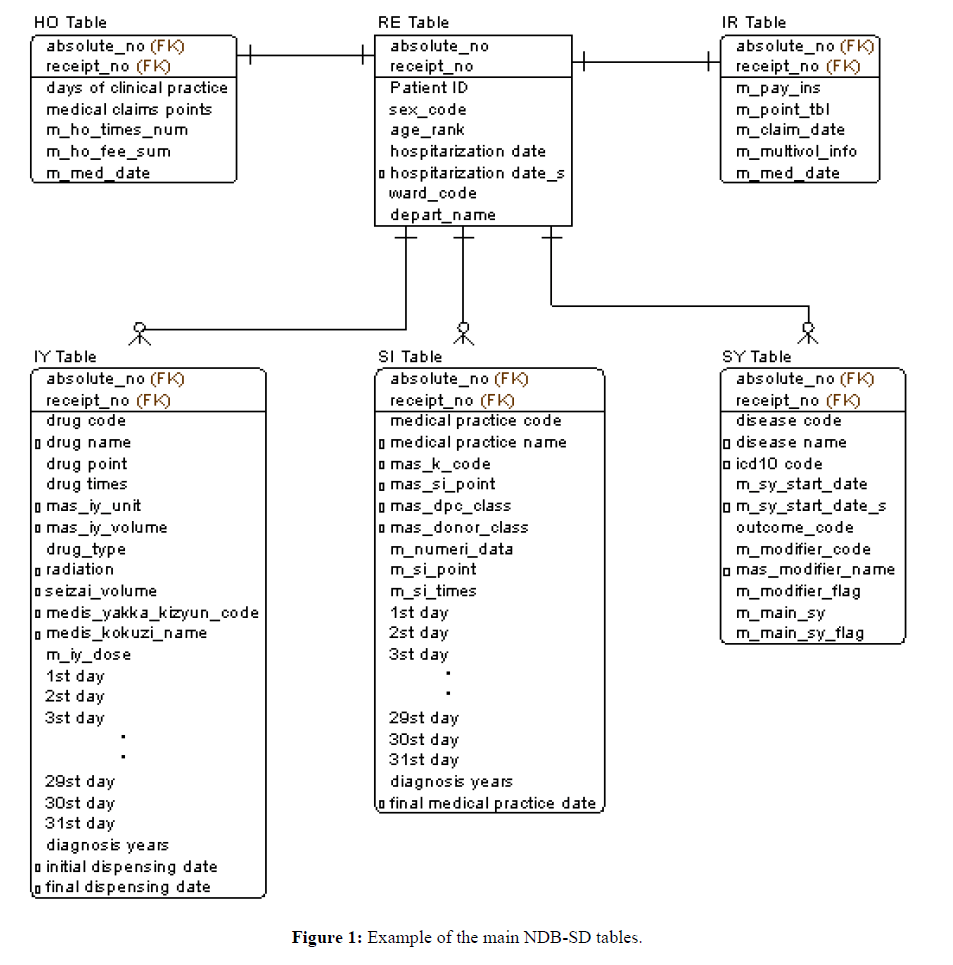
This study uses the NDB-sampling dataset (NDB-SD), which contains a random sample of insurance claims data over a 1-month period. To obtain this NDB-SD, which comprised the items “medical outpatient,” “medical inpatient,” “dispensing” and “DPC,” we collaborated with the Department of Obstetrics and Gynecology and the Primary Care and Emergency Medicine Department of Kyoto University Hospital; for the present study, we only used the data concerning “medical outpatient,” “medical inpatient” and “dispensing.” Figure 1 shows the NDB-SD tables used in this study. Figure 1 shows the healthcare claim number issued for a specific patient by an individual medical institution corresponds to (receipt_no). In the Japanese system, such claims are issued once per month. All tables are based on RE tables, which list patient details, such as age and sex, labelling them with a unique patient ID. The IR and HO tables correspond to the RE table through a 1:1 relationship with (receipt_no); the IR table stores data on medical institutions, whereas the HO table stores data associated with medical care fees. Multiple IY, SI, and SY tables, which store data on medications, medical practice, and injuries and diseases, respectively, can be linked to a single RE table. It is common for a single patient to have multiple entries regarding such information, and they can be distinguished by differentiating between (receipt_no) and (absolute_no) from one another.
Method
This paragraph contains the definitions of the terminology used in this study. In database studies, the names of the columns in tables are labelled “attributes;” therefore, this term will be used exclusively to refer to column names in database structures. The term “attributes” approximately corresponds to “variables” in datasets for biostatistics or epidemiological analyses.
One of the problems concerning data stored in the NDB is the definition of the data. For example, all dates stored in the NDB are in Japanese (gengou) calendar years and, thus, are not suitable for analysis using a statistical analysis software. Furthermore, for the analysis of drugs, there were no independent attributes for the units used in prescriptions (e.g., mg); only drug codes were provided. Therefore, to perform an efficient analysis, it is necessary to complement the data with an external master file or other means.
We began by reviewing studies that used NDB [7]. This review encompasses studies that used NDB data from the time it was first made available to general users (FY2001) to those that used data available in March 2017. In total, 105 examples of NDB use for conference presentations, reports, public surveys, and articles were examined. However, of these, only 20 involved data analysis and were published in journals; these 20 will hereafter be collectively referred to as “NDB-Literature”. Next, the tables, figures, and sentences in the NDB-literature that contained analysis results were carefully reviewed to count the data entries used or derived in these studies, which consequently ranged between a minimum of four and a maximum of 2,812. Studies that analyzed a large number of drugs which tended to use higher amounts of data. The data were then organized and grouped into 30 attributes. Some of these attributes are easily extractable from age- or sex-based databases, whereas others, such as the Charlson Comorbidity Index, a frequently used index for epidemiology studies, require extensive labor to derive because they are not stored in the current NDB-SD.
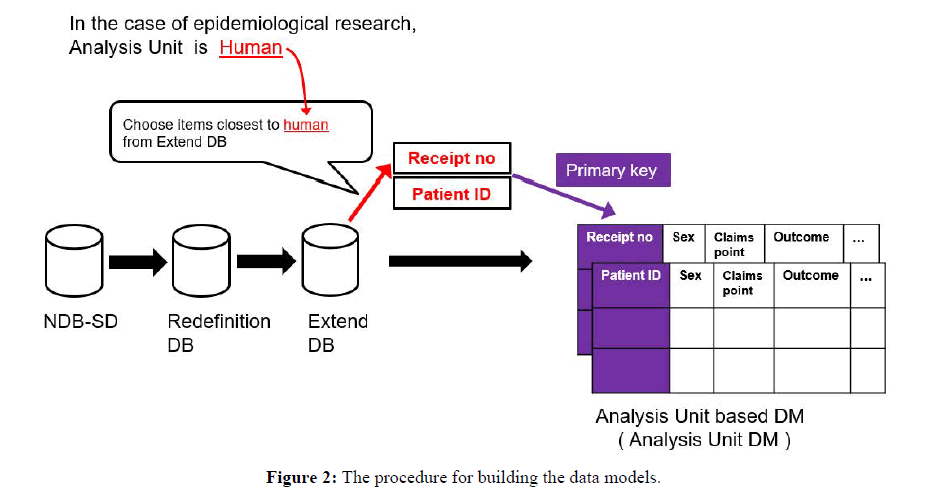
As a preliminary step (phase 1), a redefinition database (Redefinition DB) was constructed to convert or redefine attributes in the existing NDB-SD into more general definitions that could be more easily manipulated on a calculator. Next, an extended database (Extended DB) was constructed to store the attributes required for epidemiological studies (phase 2). In the final step, an analysis unit data model (Analysis Unit DM) was constructed based on the previous steps (phase 3) to resolve the various problems encountered in epidemiological analysis. The procedure for database construction is detailed below.
Phase 1: Construction of Redefinition DB
The redefinition DB was constructed based on NDB-SD, primarily comprising data-shaping date-based information. The NDB-SD contains data concerning dates, such as admission, discharge, initiation of care, death, prescriptions, and medical practice, and these data are dispersed across many tables. In the redefinition DB, all of these attributes were converted into data in Gregorian calendar format, which makes attributes dated across different gengou or calendar systems compatible for calculation using arithmetic operations hosted by a statistical analysis software or SQL. In other words, this enables the easy calculation of outcomes, such as the “number of days between the date of admission and death.
Phase 2: Construction of Extended DB
The extended DB allows attributes obtained in the NDB literature survey to be added efficiently to the Analysis Unit DM while maintaining the NDB-SD table structure. Specifically, this consists of data sharing or adding external master data to the redefinition DB. Furthermore, prescribed dosages (mg) of drugs and disease names were assigned Charlson Comorbidity Index scores, which allowed users to obtain new per-patient attributes that were converted into Charlson Comorbidity Index units. In addition, master files for “disease name,” “medical practice,” and “drug name” were obtained from MHLW [8], and the ICD10 standard disease name master files and the drug HOT code master files were obtained from MEDIS-DC [9]. Finally, the attributes obtained through these steps were added to the existing NDB SD tables. In Figure 1, the attributes in each table marked with the prefix “o” are those that were newly added to NDB-SD to develop the extended BD.
Phase 3: Construction of Analysis Unit DM
Selection of primary key:
In the relational model, unique attributes in each of the tables are called “primary keys.” As epidemiological research studies human populations, the analysis unit is “human.” The closest attributes to “human” in the extended DB are {patient ID} and {receipt no}; thus, in this study, these attributes were treated as the primary keys, and an analysis unit DM comprising one row for each analysis unit was constructed.
Selection of attributes
The table structure of the extended DB is identical to that of the conventional NDB-SD; therefore, the data of interest to the researcher are still dispersed among multiple tables. However, the extended DB has attributes that are fundamental for inclusion in the analysis unit DM. Therefore, we used SQL, a database management language, and aimed to collate the attributes obtained from the NDB-literature survey into single tables in terms of the analysis unit (patient ID, receipt no). This resulted in the implementation of 18 of the 30 attributes obtained from NDB-literature (Figure 2) to construct the per-analysis unit data model. To calculate the Charlson Comorbidity Index, we used a study that proposed codes based on the ICD10 [10] as a reference and implemented scoring of diseases that co-existed in a patient over a 1-month period.
Results
The attributes obtained from NDB literature are listed in Table 1. The “groups” of attributes in Table 1 are classified as follows: “G1” are attributes that appeared in at least 50% of studies in the NDB-literature; “G2” are those that were not as frequent, but were still common in the NDB-literature; and “G3” relates to attributes with a relatively low level of generality (G3 attributes were used in studies in the NDB-literature, but were specific to each study objective). “Frequency” refers to frequency of appearance (maximum value: 20) in the NDB-literature. The attributes included in the analysis unit DM are shown with a “o,” while those that were not included are indicated with an “x.” Furthermore, the column titled “Extended DB” shows attributes that were included in the extended DB by default. Ultimately, the G1 and G2 attributes were implemented in the analysis unit DM, and 19 of the 30 attributes were rendered.
| Group | Attribute (variable) | Analysis Unit DM | Extended DB | Frequency (category) |
|---|---|---|---|---|
| G1 | Age group | ◯ | - | 17 |
| Sex | ◯ | - | 14 | |
| G2 | Discharge date (AD format) | ◯ | - | 6 |
| Hospitalization date (AD format) | ◯ | Hospitalization date (AD format) | 5 | |
| Charlson comorbidity index | ◯ | ICD10 code | 4 | |
| Days receiving clinical practice | ◯ | - | 4 | |
| Presence or absence of medical treatment | ◯ | - | 2 | |
| Administering of blood products for transfusion | ◯ | Type of blood products used | 2 | |
| Date of initial dispensing of drugs (AD format) | ◯ | Date of dispensing of drugs (AD format) | 2 | |
| Application of internal medicine | ◯ | - | 2 | |
| Death | ◯ | - | 1 | |
| Application of medication | ◯ | - | 1 | |
| Administering of injection | ◯ | - | 1 | |
| Application of prescription | ◯ | - | 1 | |
| Administering of inspection | ◯ | - | 1 | |
| Application of external medicine | ◯ | - | 1 | |
| Date of final dispensing of drugs (AD format) | ◯ | Date of final dispensing of drugs (A.D format) | 1 | |
| Date of final application of medical practice (AD format) | ◯ | Date of final application of medical practice (A.D format) | 1 | |
| G3 | Drug name(s) specific to the research | × | Drug name | 14 |
| Name of medical practice(s) specific to the research | × | Medical practice name | 12 | |
| ICD10 code(s) specific to the research | × | ICD10 code | 10 | |
| Prescription date specific to the research (AD format) | × | Prescription date (AD format) | 8 | |
| Disease name(s) specific to the research | × | Disease name | 7 | |
| Date of administering medical practice(s) specific to the research | × | Medical practice date (AD format) | 6 | |
| Date of commencement of diagnosis of a disease specific to the research | × | Diagnosis commencement date (AD format) | 4 | |
| Volume of medication specific to the research | × | Volume of medication (unit: mg. ml) | 4 | |
| Classification by efficacy of drugs specific to the research | × | Classification drugs by efficacy | 4 | |
| Number of administrations of medical practice(s) specific to the research | × | Medical practice name | 2 | |
| Health insurance claims points | × | - | 1 | |
| Drug’s YJ code | × | Drug’s YJ code | 1 |
Table 1: Data items obtained from the NDB-Literature.
Next, a study was conducted regarding postpartum hemorrhage (PPH) in the field of obstetrics and gynecology using the DW that was developed in this study, and an article was written based on the obtained findings [11]. This study reports changes in treatment methods and incidence of PPH. Table 2 shows how comprehensively analysis unit DM covered the variables used in the study’s analysis. Overall, six attributes were required, four of which were included in the analysis unit DM. Similarly, an epidemiological study on patients after cardiopulmonary resuscitation in emergency fields required seven attributes, of which four were covered (Table 3). Approximately 20,000 steps that were required to make a DW from the CSV file provided by NDB-SD were automated using Windows MS-DOS commands and SQL. Because these byproducts do not contain personal information, we are considering making this automated process available at no cost, including technical explanations, depending on demand.
| Data item used in the paper | Attribute (variable) | Covered |
|---|---|---|
| receipt_no | (analysis unit) | - |
| Sex | Sex | ◯ |
| Age group | Age class | ◯ |
| Death | Death | ◯ |
| Uterine atony | Disease names specific to the research | × |
| Placental abruption | ||
| Placenta previa | ||
| Retained placenta | ||
| Placenta accreta | ||
| Cervical laceration | ||
| Vaginal hematoma | ||
| Multiple pregnancy | ||
| Low-lying placenta | ||
| Uterine rupture | ||
| Placenta previa accreta | ||
| Uterine inversion | ||
| Amniotic fluid embolism | ||
| Volume of RCC (ml) | Volume of medication (mg) | ◯ |
| Volume of FFP (ml) | ||
| Volume of PC (ml) | ||
| Cesarean delivery | Medical practice specific to the research | × |
| Operative vaginal delivery | ||
| Surgical intervention | ||
| Intrauterine balloon tamponade | ||
| Arterial embolization | ||
| Hysterectomy |
Table 2: How comprehensively the attributes required for an epidemiological research of PPH, which was previously published in the Journal of Maternal-Fetal & Neonatal Medicine (DOI: 10.1080/14767058.2018.1465921), were covered by the Extended DB.
Discussion
This study involved adding attributes frequently used in epidemiological research to the NDB-SD to construct the extended DB, and then, based on that, constructing the analysis unit DM.
First, we discuss the extent to which the attributes required for actual epidemiological research, which were identified from the NDB -literature survey, were covered by the analysis unit DM. Examining epidemiological studies in the areas of obstetrics, gynecology, and emergency care, over 50% of the attributes actually used in the studies were covered by attributes that were implemented in the analysis unit DM. The main reason for this was that the G1 attributes and several G2 attributes in the analysis unit DM (Table 1) were ultimately used in the actual studies. Unlike attributes, such as age group and sex, which are almost always used in epidemiological studies, the use of G2 attributes is highly likely to depend on the type of database used for the study. Therefore, it is important to determine the types of attributes required by reviewing published articles in studies that have used similar databases.
As shown in Table 3, G3 attributes primarily comprised disease names, drug names, and medical practices. From the NDBliterature survey, these data items could be numerous in some studies and also tended to be specific. For example, studies that focused on many types of drugs used a large number of drug codes. However, when data with lower generalizability were included in the analysis unit DM, the browsability of the DW was decreased; therefore, it is considered more suitable for users to add such data as attributes to the analysis unit DM as needed. For this reason, G3 attributes were not included in the analysis unit DM.
| Data item used in the paper | Attribute (variable) | Covered |
|---|---|---|
| receipt_no | (analysis unit) | - |
| Sex | Sex | ◯ |
| Age group | Age group | ◯ |
| Death | Presence or absence of death | ◯ |
| Administering of blood products for transfusion | Administering of blood products for transfusion | ◯ |
| Postresuscitation encephalopathy | Disease specific to the research | × |
| Adrenalin | Drug specific to the research | × |
| Closed chest cardiac massage | Medical practice specific to the research | × |
| Tracheal intubation | ||
| Artificial respiration | ||
| Defibrillation | ||
| EEG | ||
| CT | ||
| Induced Hypothermia | ||
| CAG | ||
| PCI | ||
| Hemodialysis | ||
| PCPS | ||
| Medical expenses for first aid | ||
| Medical expenses for intensive care | ||
| Postresuscitation encephalopathy |
Table 3: How comprehensively the attributes required for epidemiological research of patients after cardiopulmonary resuscitation, which was previously presented at a meeting of the Japanese Association for Acute Medicine, were covered by the Extended DB.
Technical improvements achieved through the present study made it possible for all the G1, G2, and G3 attributes shown in Table 3 to be added using a query (SQL) in patterns of approximately 15 rows. For example, when considering adding “ICD10 code specific to the research,” a G3 attribute, to the analysis unit DM, ICD10 codes are already assigned to all disease names in the extended DB; thus, because the extended DB is equipped with fundamental attributes by default, it is relatively easy to add new attributes to the analysis unit DM [11].
Thus, the analysis unit DM constructed using this study’s DW provided all frequently used attributes and also made it relatively easy to add other attributes to increase the efficiency of epidemiological analysis for general clinicians. In addition, it was a new discovery that we could cover a considerable number of attributes in a newly conducted epidemiological study with attributes obtained from NDB-literature, which includes only 20 documents. From this fact, it can be said that the attributes that are necessary for epidemiological research are likely to be patterned.
Finally, this study has some limitations. First, the NDB-SD used in the present study comprises sample data for a period of 1 month; thus, it is only suitable for cross-sectional research. We plan to obtain health insurance claims data suitable for cohort studies to check whether the methods in this study can be applied to such studies.
Conclusion
This study focused on identifying the attributes that are frequently used in epidemiological analyses and constructing a DW that facilitates the analysis of such attributes. Generally, when engineers construct such DWs for epidemiological analysis, they collaborate with epidemiologists. However, epidemiology encompasses a wide range of specializations; therefore, epidemiologists’ knowledge of areas other than their area of expertise is typically limited. Therefore, it is unlikely that there will be intrinsic motivation among various specialists to exhaustively identify the attributes frequently used across the epidemiological research field. We hope that building a comprehensive DW in the present study represents the first step towards constructing an NDB that can be used across specializations.
Abbreviations
NDB: National Database of Health Insurance Claims and Specific Health Checkups of Japan
NDB-SD: NDB Sampling Dataset
DPC: Diagnosis Procedure Combination
DW: Data Warehouse
Conflicts of Interest
The authors declare no conflicts of interest with respect to this research and paper.
Acknowledgments
We are deeply grateful to the Editage proofreading team for completing this manuscript.
Funding
This work was supported by ISHIZUE 2022 of Kyoto University and AMED under Grant Number 21lk1503009j0005
References
- Codd EF. A relational model of data for large shared data banks. Commun. ACM. 1970;13(6):377-87.
- Date CJ. Database in depth: relational theory for practitioners. " O'Reilly Media, Inc."; 2005.
- Kim L, Kim JA, Kim S. A guide for the utilization of health insurance review and assessment service national patient samples. Epidemiol Health. 2014;36.
- Clinical Data Interchange Standards Consortium. Analysis data model v2.1. [cited 2022 Sep 10]; Available from: https://www.cdisc.org/standards/foundational/adam/adam-v2-1
- Okamoto K. Analysis of the receipt and health check up information database in Japan. 2016;259:755–759.
- Ministry of Health, Labour and Welfare. Website on the provision of healthcare claims data and data from specific health examinations.
- Ministry of Health, Labour and Welfare. Summary of deliverables provided by third parties.
- Ministry of Health, Labour and Welfare. Various information of medical fee.
- Medical information system development center. MEDIS standard master.
- Quan H, Sundararajan V, Halfon P. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005:43:1130–39.
- Sato M, Kondoh E, Iwao T, Hiragi S, Okamoto K, Tamura H, et al. Nationwide survey of severe postpartum hemorrhage in Japan: an exploratory study using the national database of health insurance claims. J Matern. -Fetal Neonatal Med. 2019 Nov 2;32(21):3537-42.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref